Senior Researcher Tatsuya NAKANO
Division of Safety Information on Drug, Food and Chemicals
National Institute of Health Science

The goal of this project is to develop a predictive algorithm for the in silico screening system (BioStation), including the determination of receptor-ligand binding energy and the analysis of interaction energies based on quantum mechanics. BioStation consists of the following four subsystems:
(1) Molecular interaction analysis with the ab initio FMO method (ABINIT-MP)
ABINIT-MP performs a molecular interaction analysis with the ab initio FMO method, which predicts receptor-ligand binding energy and interaction analysis based on quantum mechanics.
(2) Visualization with Java and Java3D (BioStation Viewer)
BioStation Viewer is a visualization tool for the molecular interaction analysis with Java and Java3D.
(3) Docking and virtual ligand screening (BioStation Dock, BioStation Launcher)
BioStation Dock is the in silico screening system using a specific force field for the molecular interaction and the replica-exchange Monte Carlo method, and BioStation Launcher provides the integrated environment with Java and XML.
(4) Target database (KiBank)
KiBank is the integrated database system for ligand compounds such as pharmaceuticals, nuclear and membrane receptors, enzymes, transporters, ion channels and ion pumps.
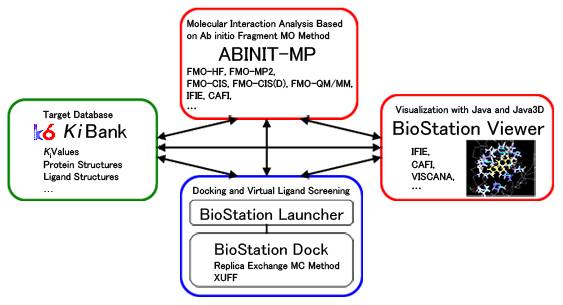
Figure Schematic diagram of the quantum molecular interaction analysis system (BioStation)



