We previously developed “ProteinDF,” the world’s largest density-functional-approach-based system for calculation of canonical molecular orbitals for proteins, a revolutionary system that enables calculation of the all-electron molecular orbitals, energies, and atomic charges of proteins having a few hundred residues.
- Home
- Research Content
- Bio, Nano Molecule Simulator
-
Bio, Nano Molecule Simulator

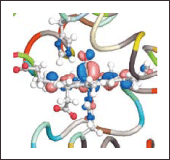
Using this system as the basis, in this project we will conduct research and development that takes into account advanced applications to enable us to come up with a cutting-edge system that can serve as a foundation for generating innovations in industry and academia. Particularly, a) to enable more direct analysis and comparison with experiments, we will develop functions for computing spectroscopic properties and other important properties.
These will accelerate the creation of an environment that inextricably links experiment and theory. b) We will develop integrated simulation functions for proteins and functional nanomolecules that contribute to the manufacture of new bio- and nano-scale materials; such simulation is only possible with huge canonical molecular orbital methods. c) Using our previous experience in all-electron calculations for metalloproteins, we will develop a system to analyze the P450 hydroxylase, the major enzyme involved in drug metabolism. Our goal is to provide an analysis platform based on first principles, unlike conventional empirical methods. d) Finally, to enable the use and reuse of the results of these systems over a wide range of applications, we will develop methods for making the huge protein wave function database available. We expect that this project will be instrumental in the construction of a full-scale virtual molecular biology laboratory.
All-Electron Wave-function Calculations of Proteins by DFT
ProteinDF

